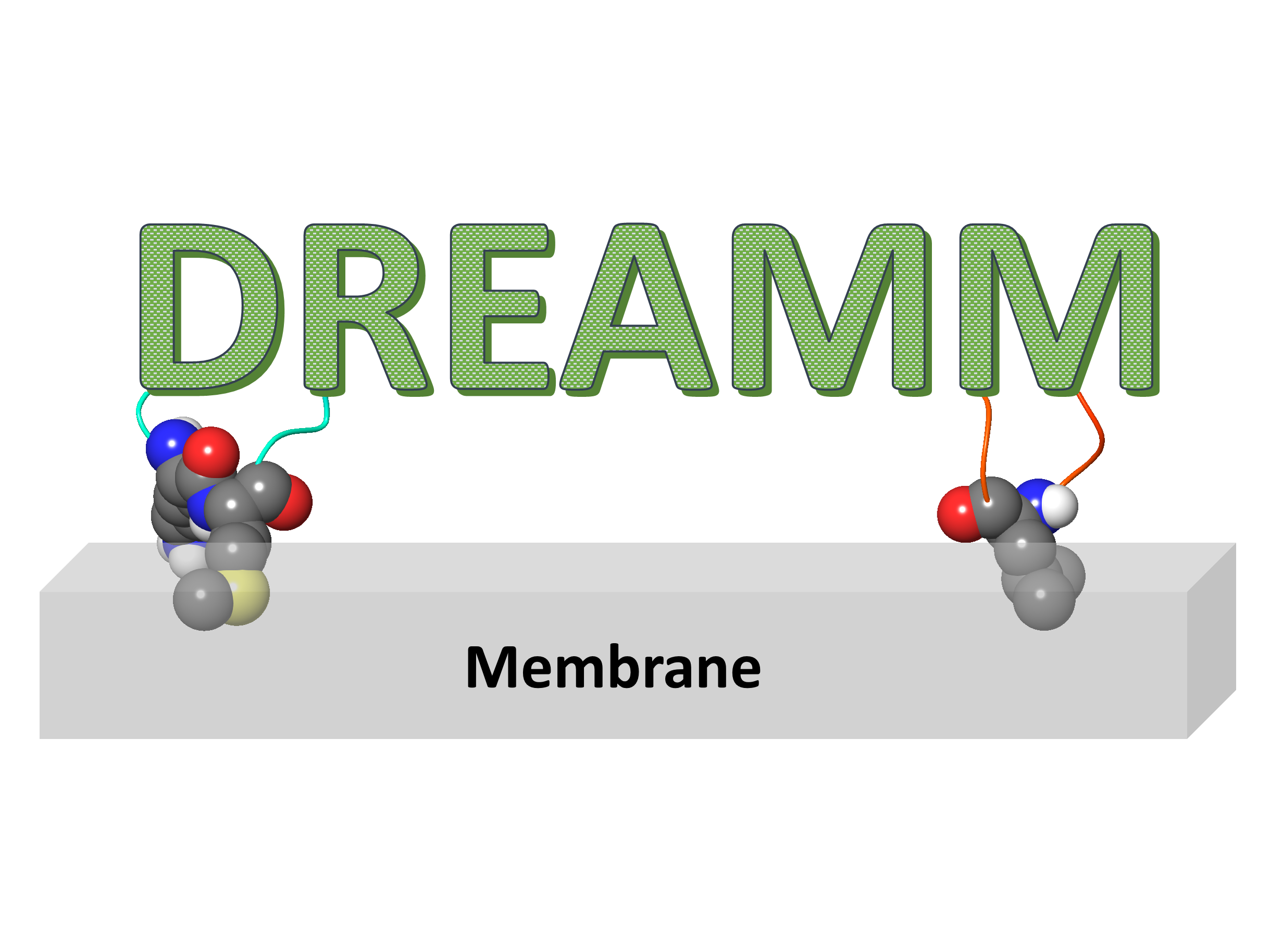
DREAMM
DREAMM is a novel machine learning tool that predicts the protein-membrane interfaces of peripheral membrane proteins.
OK
DREAMM is a novel machine learning tool that predicts the protein-membrane interfaces of peripheral membrane protein, and optionally predicts binding sites near the predicted membrane-penetrating residues in protein conformational ensembles. As an output, the user can retrieve the membrane-penetrating residues in a .csv file and if the user's choice was to predict binding sites, a .zip file will be downloaded including the abovementioned .csv file, the binding pocket predictions, the visualizations, and summarized the binding site clustering results.
Resource organization
Resource providers

Soranou Ephesiou 4, 11527 Athens, Greece (EL)

Soranou Ephesiou 4, 11527 Athens, Greece (EL)
Website
Readiness
TRL9
Domain
Subdomain
Category
Subcategory
Target users
Access type
Access mode
Tags
Contact
Zoe
Cournia,
Associate Professor
email: zcournia@bioacademy.gr
phone: +302106597195
